Work
2020 - Present
Murphy Labs, LLC
Owner of an indie software company developing web and mobile applications.

Memolli
(memolli.com)
iOS and web app for tracking products you try, such as coffee, wine, tea, and whisky. Users can capture images, detailed tasting notes, ratings, and more. Customizable review templates and product categories. Implemented offline access, syncing across devices, and AI to detect product information from images.

Strivocal
(strivocal.com)
Web app that offers voice training software. Provides real-time feedback while the user speaks: pitch, formants, volume, and gender. Uses both standard audio algorithms and AI for real-time, low latency inference on streaming audio.

Genderfluent
(genderfluentapp.com)
Web and iOS app that offers voice training software for the LGBT community. Provides real-time feedback while the user speaks: pitch, formants, volume, and gender.
2021 - 2024

Mission Bio
Staff Bioinformatics Scientist (2023 - 2024)
- Led technical development of a new assay extending the scMRD product software & algorithm (python).
- Met with potential clients to solicit feedback on product concepts (e.g. panel design)
- Extended & optimized VDJ analysis pipeline to reduce runtime by 80% (Python / nextflow)
- Partnered with product & software teams to coordinate & prioritize software sustaining work for scMRD product.
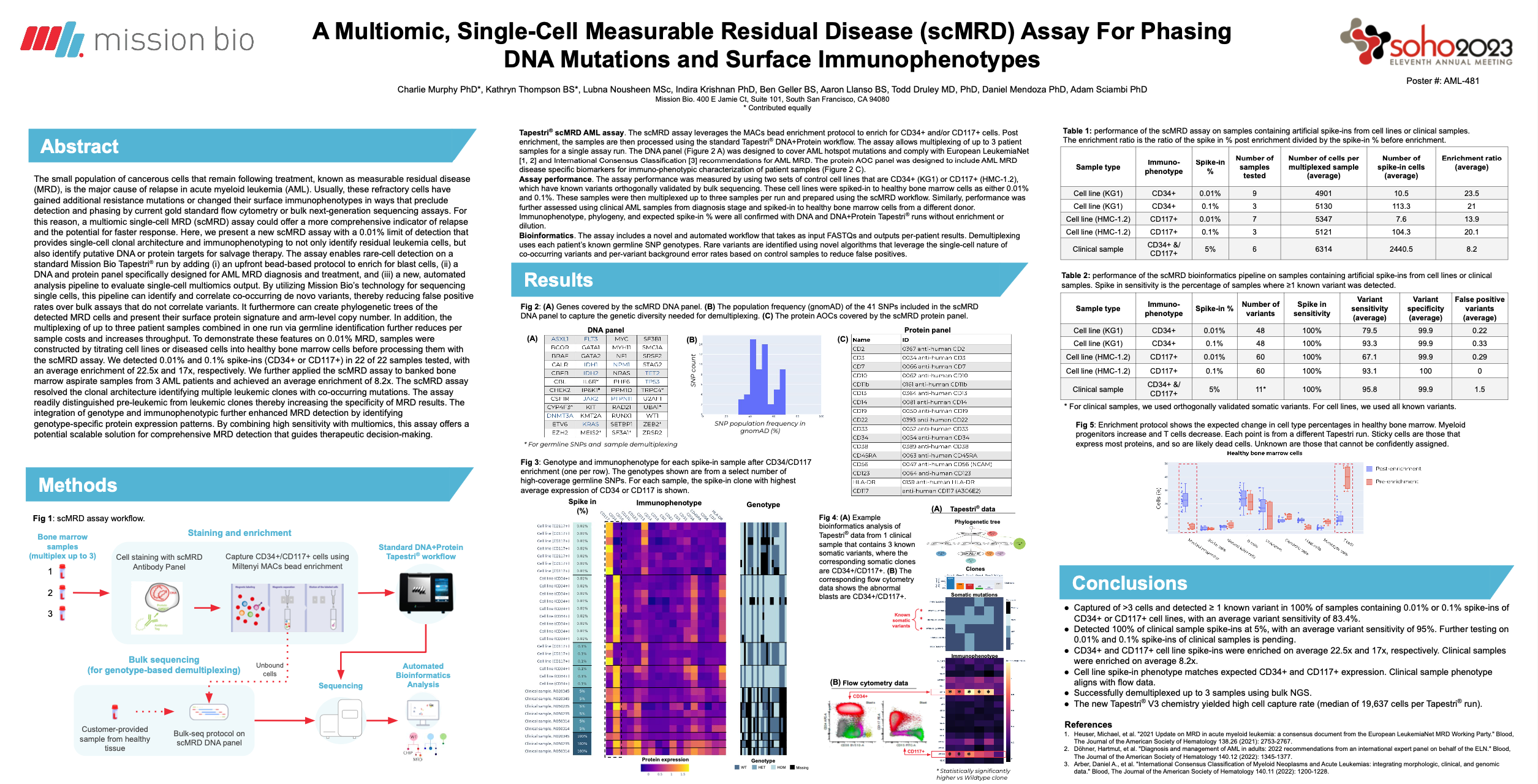
Patents:
- Sciambi, Adam and Murphy, Charles Joseph (2023). True variant identification via multianalyte and multisample correlation (Patent No. WO2023196928A3)
- My contribution was the development of a novel algorithm and code for identifying rare somatic variants in single-cell data, which used a site-specific statistical model to account for technical noise and biological variability.
Senior Bioinformatics Scientist (2021 - 2023)
- Launched an industry-first scMRD assay and software product used by nine clients.
- Led technical development of a novel data analysis pipeline & algorithms (Python/Nextflow) and interactive report (HTML/Jinja2). Partnered with software team for code testing and deployment strategies to cloud/on-prem.
- Worked closely with product & software teams to develop product & software requirements, estimate timelines, demo software concept to leadership & potential clients, and pass product development phase gates.
2020 - 2021

Memorial Sloan Kettering
Senior Computational Biologist
- Extended & optimized a cfDNA data analysis pipeline (Python, CWL) that processed tens of GB of data / sample.
- Ported the pipeline from a local compute cluster to AWS cloud by wrapping tools in Docker containers.
- Built scalable tool to identify sample swaps/contamination and output interactive charts (Python / Plotly).
- Led a collaborative analysis of two cfDNA datasets to identify novel clinical applications of the assay.
- Reviewed sample QC and mutations from MSK-ACCESS datasets before handoff to other teams for analysis.
2018 - 2020

Basepair
Senior Bioinformatics Scientist
- Led technical development of 13 new analysis pipelines (Python) and report generation that were deployed to an AWS cloud environment, as well as supervising an intern.
- Partnered with full stack developers to reviewed code to create front-end visualization of pipeline results (React).
- Improved backend pipeline system, such as adding an automated error catching & ticket creation system on pipeline failure, logging of pipeline runtime metrics (Python, Django, and AWS).
- Managed 14 consulting analysis projects for external clients, including several enterprise clients to prototype & maintain bioinformatics pipelines for their products (e.g. TCR/BCR repertoire analysis, CUT&RUN/Cut&Tag).
- Facilitated user retention/growth via webinars, workshops, blog posts, and technical support (calls/tickets).
2013 - 2018

Cornell University
PhD Student in Computational Biology
- Authored data analysis workflow to find tumor-specific therapeutic targets (Nextflow/Python/R)
- Collaborated with over a dozen other scientists to analyze their data using standard or custom bioinformatics pipelines and bespoke statistical analyses; resulting in 12 peer-reviewed publications.
2008 - 2013

University of Wisconsin - Milwaukee
Projects
2025 - present
uplot-react-native
React native wrapper around uPlot. Works on web, iOS, and Android. Wraps uPlot in a WebView on iOS and Android.
github.com/murphycj/uplot-react-native
2018 - 2021
AGFusion
Free web app and python package to annotate gene fusions. Cited by 22 publications. (Python, Javascript/React, AWS)
React-based webapp:
github.com/murphycj/agfusionweb-react
Python package:
github.com/murphycj/AGFusion
