Written by: Charlie Murphy
Introducting a webapp for annotating and visualizing gene fusions
Check it out here: https://www.agfusion.app/
What is it for?
Gene fusions are a type of genomic mutation that is commonly associated with different diseases such as cancer [1]. As you likely know, the human genome has tens of thousands of genes spread across its 46 chromosomes. However, mutations can occur that “rearrange” parts of these chromosomes, which sometimes result in the “merging” together of two genes. One important impact of this is that sometimes this new compound gene produces a new protein product, or alternatively, it can eliminate the functionality of one or both genes. Either way, these types of mutations can either cause the disease or help it progress.
I previously made AGFusion, a Python package (https://github.com/murphycj/AGFusion) to visualize and annotate gene fusions [2]. However, since using the software requires programming ability, I wanted to introduce a web-based and user-friendly application for non-programmers. The new web application is essentially independent of the AGFusion Python package, with all the core code rewritten in javascript. However, both applications have all the same capabilities. Below I highlight the key features of my new web application.
Features
Input parameters
The software only needs the following pieces of data per fusion:
- 5’ gene partner
- 5’ gene partner junction location
- 3’ gene partner
- 3’ gene partner junction location
- The reference genome used to call fusions (human and mouse is supported)
Alternatively, you can provide either a generic TSV/CSV-formatted file or the output file from one of the supported fusion finding-algorithms (see below).
Output
Prediction
- Functional effect of the gene fusion (e.g. in-frame, out-of-frame, etc.)
- Annotates proteins with domains (e.g. PFAM database and others).
Downloadable data
- FASTA files of cDNA, CDS, and protein sequences.
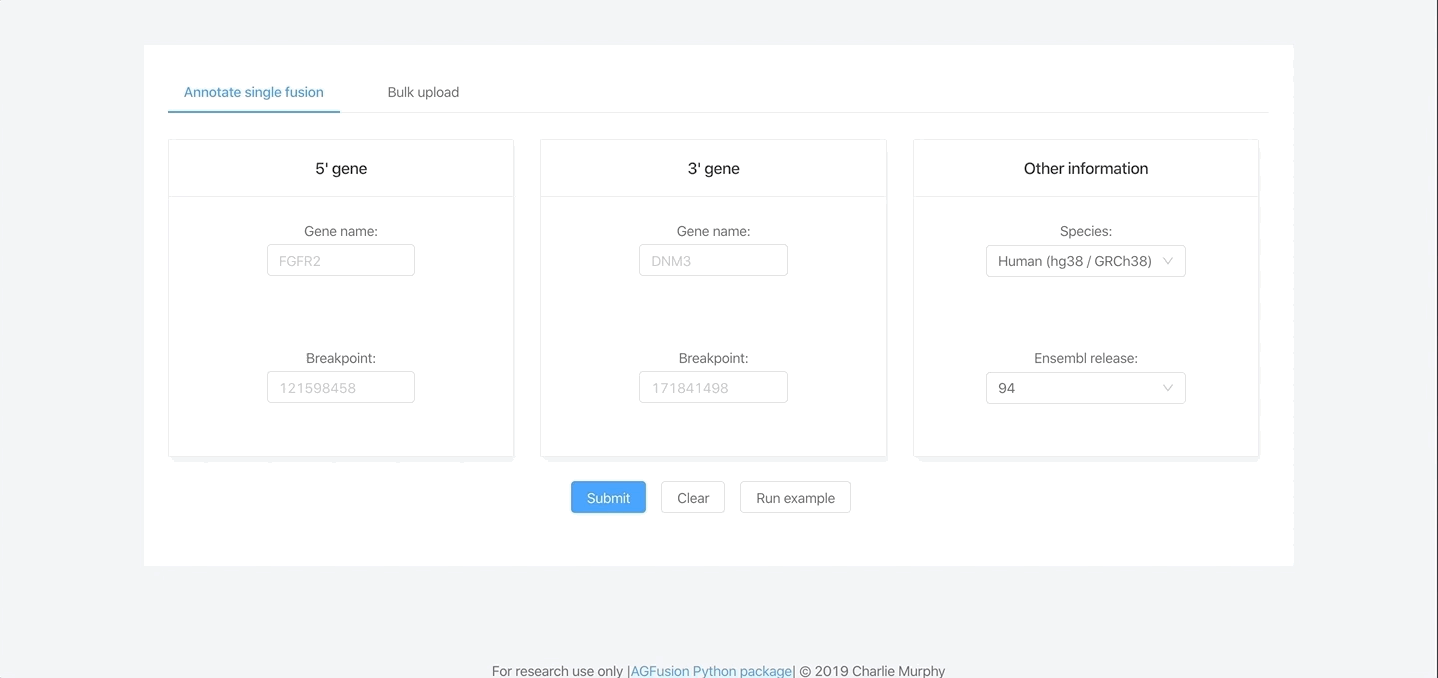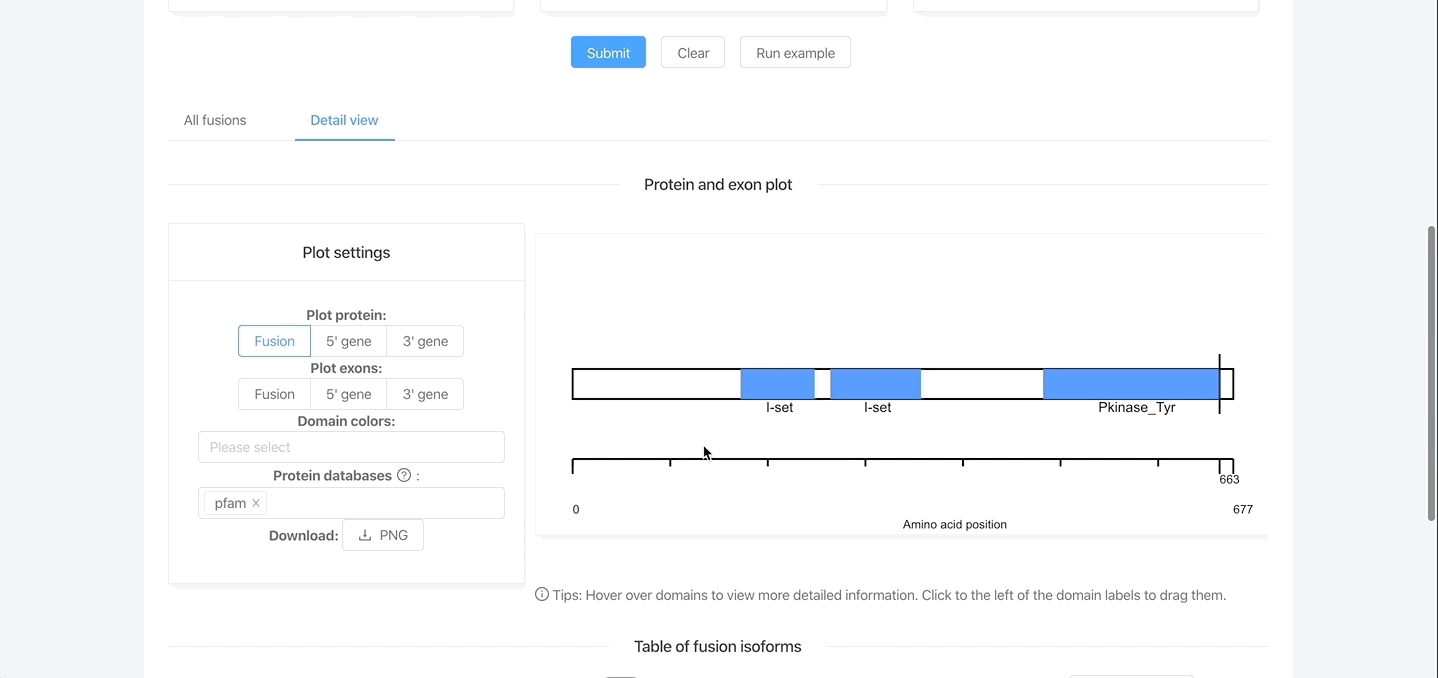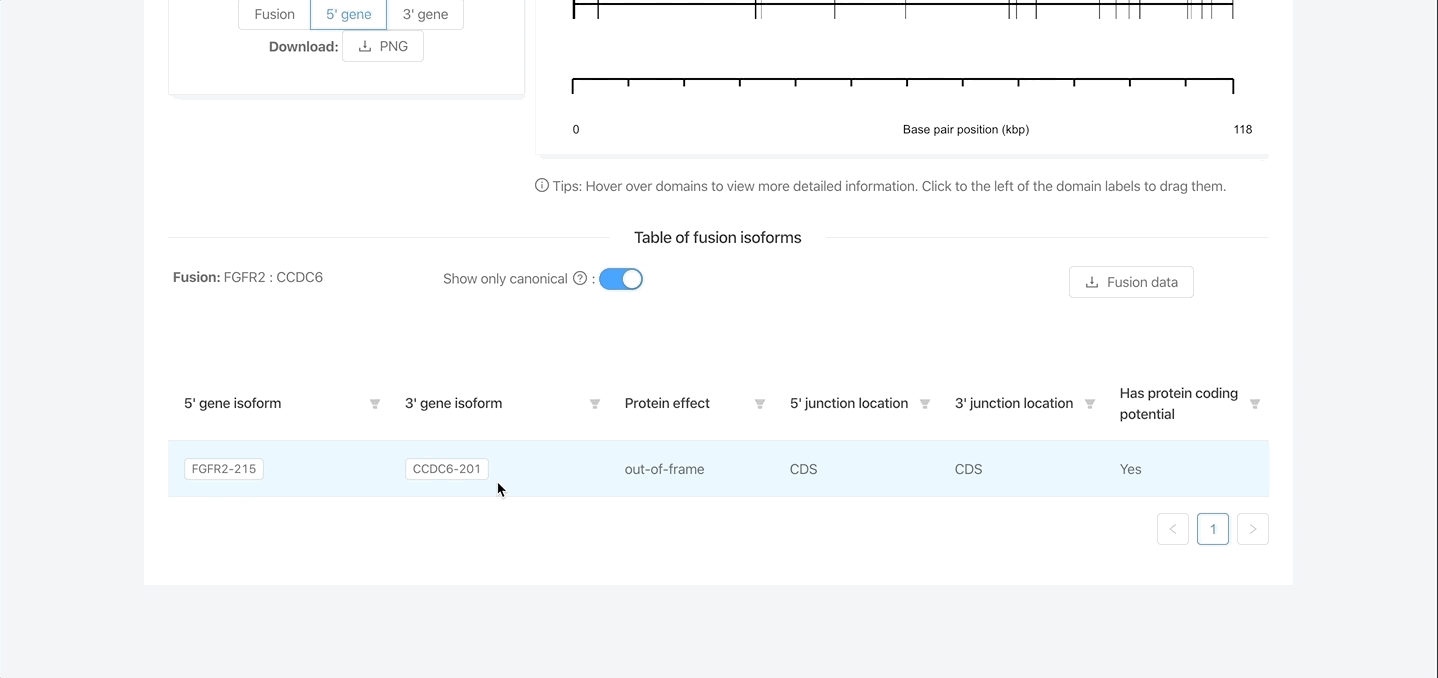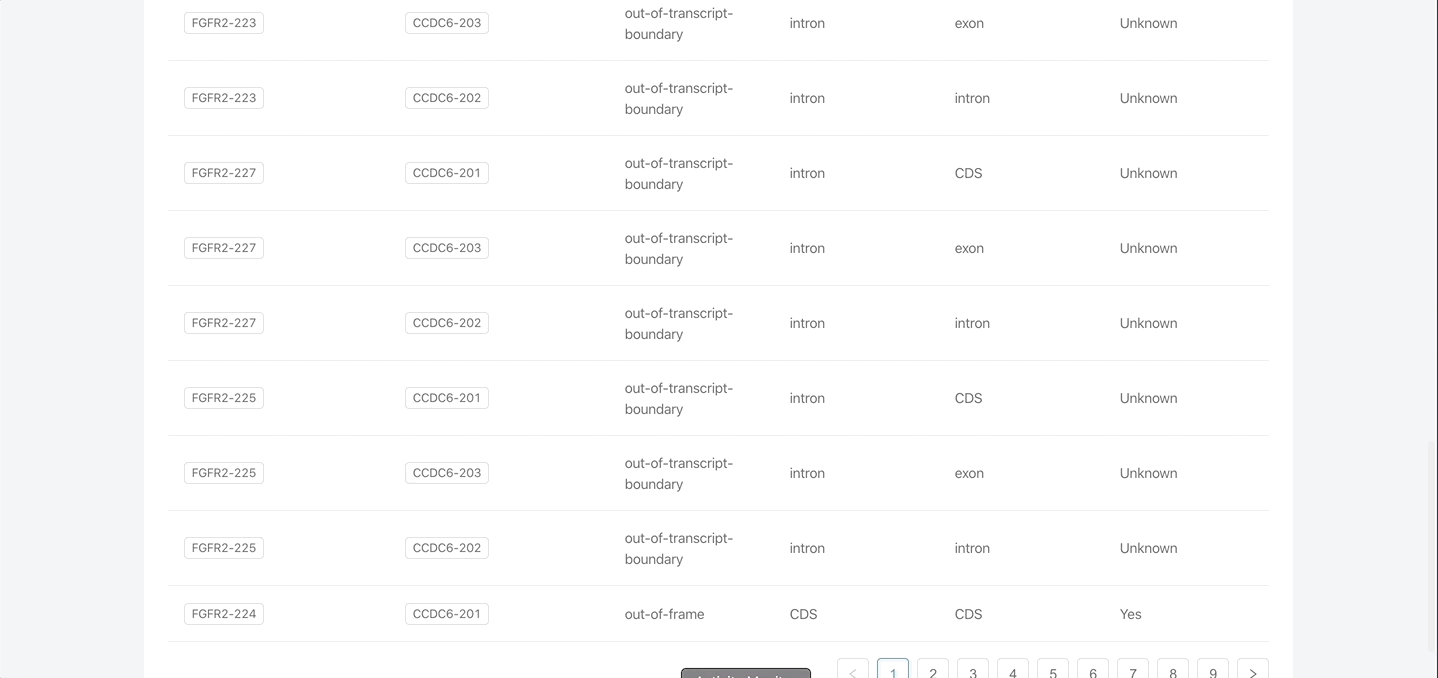
- Plots of protein domain and exon architectures.
- Various tables containing many additional details about the fusion(s), including protein domain data, exon structures, etc.
Misc.
- Data and plots on exon structure and protein domain of the wild-type version of the fusion gene partners.
- Provides data for all isoform combinations of the fusion.
Interactive figures and tables
- Table 1: All fusions
- Table 2: Detail view
- Protein/exon figures
Supports output files from fusion-finding tools
The following fusion-finding algorithms are supported:
- Bellerophontes
- Chimerascan
- ChimeRScope
- DeFuse
- EricScript
- FusionCatcher
- FusionInspector
- FusionMap
- JAFFA
- MapSplice
- STARFusion
- TopHatFusion
If your fusion-finding algorithm is not supported, you can either ask me to add support or just format it into a simple CSV or TSV file. Each row in the file should contain one fusion like so: gene1,gene1Junction,gene2,gene2Junction (e.g. FGFR2,1231231,DNM3,522312).
Technical
In case you are curious, this application is completely serverless and uses the following technologies:
- Frontend: React
- Data storage: AWS DynamoDB
- Site served from AWS S3.
If you want to browse or contribute to the code, go here: https://github.com/murphycj/agfusionweb-react
References
- Maher, Christopher A., et al. “Transcriptome sequencing to detect gene fusions in cancer.” Nature 458.7234 (2009): 97.
- Murphy, Charlie, and Olivier Elemento. “AGFusion: annotate and visualize gene fusions.” bioRxiv (2016): 080903.
